Deep learning for NeuroImaging in Python.
Note
This page is a reference documentation. It only explains the class signature, and not how to use it. Please refer to the gallery for the big picture.
- class nidl.volume.transforms.preprocessing.spatial.resample.Resample(target: float | tuple[float, float, float] = 1.0, interpolation: str = 'linear', **kwargs)[source]¶
Bases:
VolumeTransformResample a 3d volume to a different physical space.
This transformation resamples a 3d (or 4d with channels) volume to a new spacing, effectively changing its shape. It uses a provided RAS-affine matrix to interpret voxel coordinates in physical space. Check Nibabel documentation on image orientation.
Internally, it uses SimpleITK for fast and robust resampling.
It handles
np.ndarrayortorch.Tensoras input and returns a consistent output (same type).Input shape must be
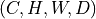 or
or 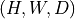 .
.- Parameters:
target : float or tuple of floats, default=1
Output spacing
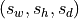 in mm. If only one value
in mm. If only one value
 is specified, then
is specified, then 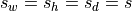 .
.interpolation : str in {‘nearest’, ‘linear’, ‘bspline’, ‘cubic’, ‘gaussian’, ‘label_gaussian’, ‘hamming’, ‘cosine’, ‘welch’, ‘lanczos’, ‘blackman’}, default=’linear’
Interpolation techniques available in ITK. linear, the default in nidl for scalar images, offers a good compromise between image quality and speed and is a solid choice for data augmentation during training. Methods such as bspline or lanczos produce high-quality results but are slower and best used during offline preprocessing. nearest is very fast but gives poorer results for scalar images; however, it is the default for label maps, as it preserves categorical values. For a full comparison of interpolation methods, see [R45]. Descriptions of available methods:
nearest: Nearest-neighbor interpolation.
linear: Linear interpolation.
bspline: B-spline of order 3 (cubic).
cubic: Alias for bspline.
gaussian: Gaussian interpolation (
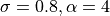 ).
).label_gaussian: Gaussian interpolation for label maps (
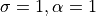 ).
).hamming: Hamming-windowed sinc kernel.
cosine: Cosine-windowed sinc kernel.
welch: Welch-windowed sinc kernel.
lanczos: Lanczos-windowed sinc kernel.
blackman: Blackman-windowed sinc kernel.
**kwargs : dict
Keyword arguments given to
nidl.transforms.Transform.
References
Examples
>>> import numpy as np >>> from nidl.volume.transforms.preprocessing.spatial import Resample >>> # Create a dummy 3D image (e.g., shape: (128, 128, 128)) >>> image = np.random.rand(128, 128, 128).astype(np.float32) >>> # Assume identity affine (voxel size 1mm in RAS) >>> affine = np.eye(4) >>> # Instantiate the transform to resample to 2mm isotropic >>> resampler = Resample(target=2.0, interpolation='linear') >>> # Apply the transform >>> resampled = resampler(image, affine) >>> print(resampled.shape) (64, 64, 64) >>> # Works the same with torch.Tensor >>> import torch >>> image_torch = torch.from_numpy(image) >>> resampled_torch = resampler(image_torch, affine)
- apply_transform(data: ndarray | Tensor, affine: ndarray | None = None) ndarray | Tensor[source]¶
Resample the input data.
- Parameters:
data : np.ndarray or torch.Tensor
The input data with shape
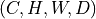 or
or 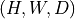
affine : np.ndarray of shape (4, 4) or None, default=None
Affine transformation matrix of the input data in RAS format defining spacing/origin/direction of the input image (in mm). This is typically given by Nibabel in this format. If None, the identity matrix is used, assuming 1mm isotropic input spacing.
- Returns:
data : np.ndarray or torch.Tensor
Resampled data with shape
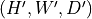 or
or
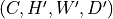 and same type as input with
and same type as input with
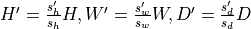 where
where 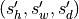 and
and 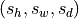 are input and output spacing (in mm)
respectively.
are input and output spacing (in mm)
respectively.
- static as_sitk(data: ndarray, affine: ndarray) Image[source]¶
Convert the input data to a SimpleITK image.
Follow us
